Computer systems have always relied on encoding information to function. From Morse code to binary encoding to the analog-to-digital encoding that enables telephone communications, encoding is embedded in our everyday technologies.
The AI revolution brought new dimensions to encoding. In 2018, Google introduced BERT, transforming how machines understand language. Today, we recognize three primary types of Language Models:
- Encoder-only models like BERT excel at understanding context but not generating text
- Encoder-decoder models like Google's T5 handle two-way tasks such as translation
- Decoder-only models, made famous by ChatGPT, predict with remarkable accuracy which words should follow in any sentence
All these models encode their inputs into tokens using various approaches.
However, at Clinithink, we've developed a fundamentally different approach.
While conventional natural language processor (NLP) systems and LLMs tokenize and vectorize language, we use a clinical NLP—specifically SNOMED CT—to structure and populate our token library. Our encoding process differentiates us:
- We break text into tagged chunks using various linguistic constructs
- Multiple parallel tasks operate on each chunk, completing in milliseconds
- The input is "mapped" to the SNOMED CT structure, with real data populating the nodes
- We qualify each concept with properties (post-coordination), resulting in millions of individual records
This approach achieves remarkable efficiency—our benchmark is one million documents per hour on a 16-core server running 32 tasks/threads.
The Power of Taxonomic Structure
The key to our speed lies in SNOMED CT's acyclic taxonomic structure. This allows us to quickly find and categorize information in a hierarchical manner. For example, a viral pneumonia is classified as:
- A viral pneumonia
- Which is an infectious pneumonia
- Which is a pneumonia
- Which is a lung disease
SNOMED CT is a poly-hierarchy (sometimes called multi-nodal), meaning concepts can have multiple parents. For instance, viral pneumonia is both a pneumonia and an infectious disease:
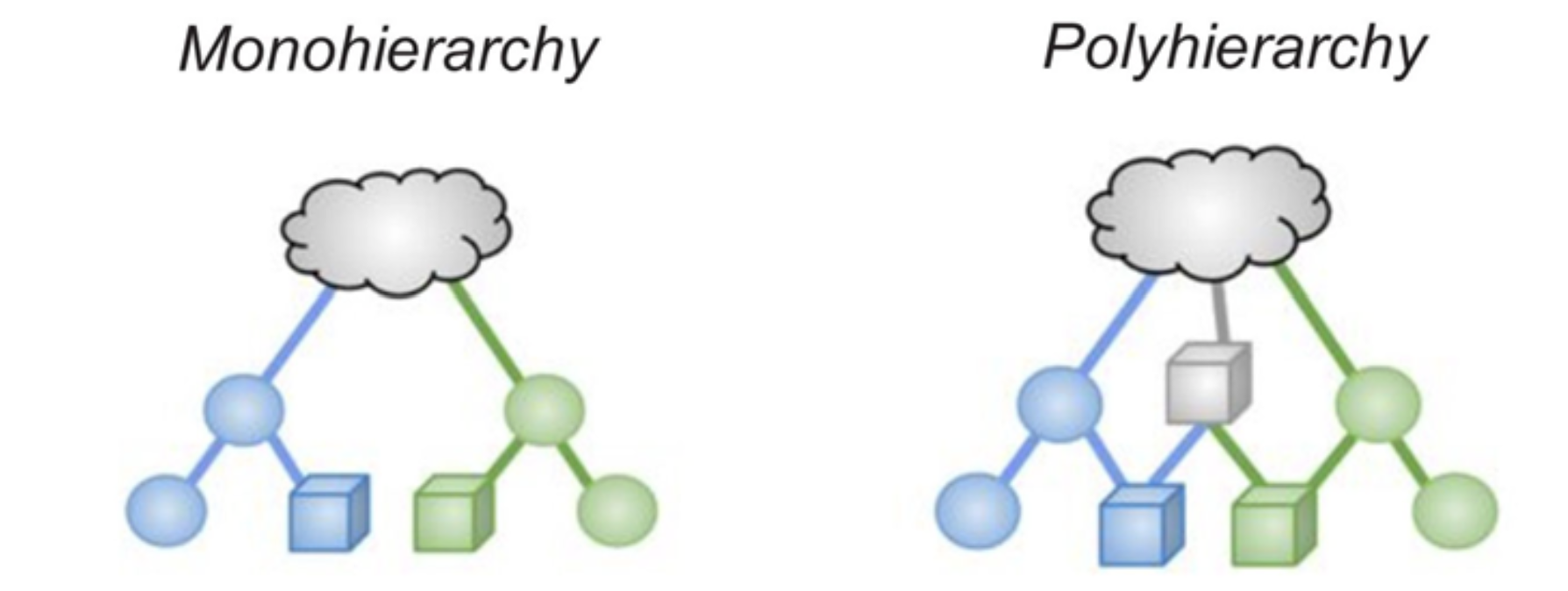
And “acyclic” here means no more than “is a” - a pictorial rendition showing the “poly” nature of the structure is shown below (i.e. viral pneumonia “is a” infectious disease).
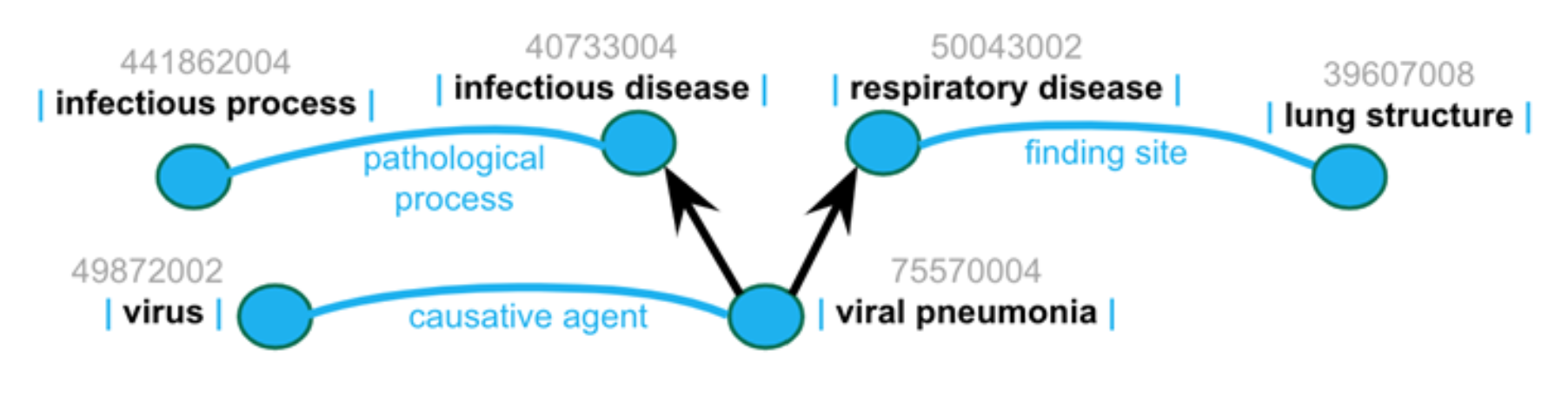
This simplified view only hints at the complexity—infectious disease has 99 children, pneumonia has 31 children (one being viral pneumonia, which itself has 15 children).
Querying: Precision at Speed
When you ask a question of an LLM, it tokenizes your words, weighs their importance, and generates probabilities from its training data to formulate a response token by token.
Our CNLP approach is fundamentally different. Because we encode input following SNOMED CT's structure, we can query with remarkable precision and speed for use cases ranging from rare disease research to early cancer detection. For example, we can instantly find all instances of "infectious disease" and its children across an entire corpus.
This structure supports complex queries, such as:
"Find all patients with cognitive decline OR mild cognitive impairment OR other memory loss AND personality change OR verbal aggression OR physical aggression OR ADLs OR ACE III score 55 to 80 OR brain atrophy OR delirium OR delirious OR family history of dementia AND NOT dementia OR severe depression OR major depressive disorder OR anxiety OR psychosis OR suicidal thoughts."
Using our interactive querying capability, we can interrogate a 30GB corpus with sub-second response times, with real world evidence published in ASCO. Our prototyped version achieves similar results with 300GB, and we've extrapolated this performance to a 3TB corpus.
Encode Once, Ask Many Times
The fundamental advantage of this approach is capturing an objective, context-free encoding of documents. This means:
- You don't need to re-encode documents to ask new questions
- Users can ask unlimited questions or drill down into specific information groups
- Multiple abstractions can be run and re-run against the same encoding
This combination of SNOMED CT's taxonomic structure and our optimized storage and retrieval methods allows us to deliver clinical insights with unprecedented speed and precision—enabling healthcare professionals to find the needle in the haystack when it matters most.
Ready to learn more?
At Clinithink, we believe healthcare AI should be both fast and verifiable. We’ve designed our CLiX engine to deliver robust clinical data extraction and coding at scale, powered by SNOMED CT and enhanced by a dynamic attention mechanism built for the real-world demands of medicine.
- Want to see it in action? Contact us to learn how Clinithink can transform your clinical data strategy.
- Curious about why tokenization matters? Check out our blog post on the role of tokenization in clinical AI.
- Looking for a deeper dive? Download our latest white paper, “Not All Healthcare AI is Created Equal,” for an in-depth look at how ontology-driven approaches outperform purely predictive models in today’s complex clinical landscape.
By bridging the gap between raw language and domain-specific context, we offer a proven way to cut through the noise—and we believe that’s exactly what healthcare needs most right now.


